PLOS Taxonomy Manager Rachel Drysdale. Image credit: Public Library of Science
At the time, the progress of genetics was painstaking, with new genes discovered one by one and their functional annotation, at least in flies, essentially binary: this is what happens when the gene works, and this is what happens when it’s turned off. But Drysdale happened to be heading the curation of a genetic database at just the time that genomics was pushed into overdrive. In 2000, the entire genome of D. melanogaster was sequenced for the first time. Geneticists flocked to fly models, and both the volume and depth of data sucked into FlyBase forced its curators to adopt a big data outlook and new computational methods.
FlyBase was also Drysdale’s introduction to working in open access science. She became involved in the Gene Ontology Project and the Phenotype Ontology Project, and when the Public Library of Science launched a new journal, PLOS ONE, that would publish every scientifically rigorous paper submitted, regardless of perceived importance, Drysdale was brought on as a Consulting Editor to help build an editorial board. Today she has over a decade of experience in the management of open access data – a field that is only around a decade old itself – and serves as Taxonomy Manager for the entire PLOS corpus.
The PLOS Taxonomy consists of a thesaurus – the set of more than 10,000 terms and synonyms that are used as subject matter tags for PLOS articles – and the complex hierarchy by which those terms are organized. The whole system had to be custom-built for PLOS, thanks to the one unruly journal that now contains roughly 80% of all PLOS articles.
“PLOS ONE has no limitation to the scope of articles that it takes,” Drysdale told Bio-IT World. “So the thesaurus that we use needs to be able to cover everything: ecological articles, surgical articles, economics articles, social science, psychology, geology, geography, computational science – across the whole spectrum.” No other scientific journal covers both the breadth and depth of PLOS ONE’s subject matter, a state of affairs that leads its taxonomy curators into all kinds of traps and contradictions.
That wasn’t always appreciated. When Drysdale was hired to manage the PLOS taxonomy in 2012, the thesaurus was going through a complete overhaul. The editors had realized that its current 3,000 terms were inadequate to the task of helping readers find relevant information amid the roughly 2,000 new articles published every month. Articles at that time were tagged by their own authors, who knew their work best but were barely acquainted with the PLOS thesaurus. Despite the limited size of the thesaurus, around a third of the terms in it had never been used – while others were so broad they could tag up to 5% of the entire published corpus, making them all but useless for searches.
In 2012, the Public Library of Science contracted with the database construction firm Access Innovations (AI) to review the taxonomy and recommend changes. AI’s engineers methodically tallied the usage of each term, flagged articles that were under-categorized, and added thousands more terms from the company’s own scientific dictionary that seemed to close gaps in coverage. PLOS and AI representatives met every week to discuss topics like which terms to add and jettison, how many nesting levels should exist in the hierarchy, and which subjects should make up the taxonomy’s highest tier.
AI also brought to the table a program called Data Harmony MAIstro (for Machine-Aided Indexing), which took over the job of assigning terms to individual articles. MAIstro is a text mining program that searches for phrases within the papers submitted to PLOS, and relates them to the terms in the thesaurus. This sounds like just about the simplest application of text mining around – until you realize that the scope of PLOS ONE is so large that the word “evolution” in an article could refer to anything from changing allele frequencies, to cultural evolution, to stellar evolution: subjects so different that they diverge at the top tier of the PLOS taxonomy. MAIstro has to be calibrated sensitively enough that these articles can, without any manual intervention, be tagged with genome evolution, celestial objects, and cultural anthropology, respectively.
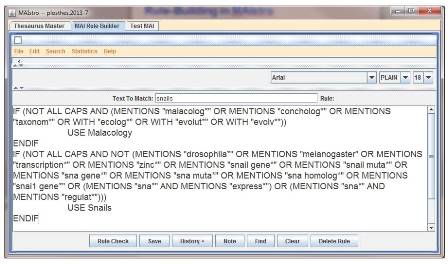
To accomplish this, Drysdale builds rules in MAIstro that respond to the context in which words appear. “There’s a massive spectrum all the way from the very simple rules to the most complicated and conditional rules,” she says. One of her favorite examples is the word “snail,” which isn’t an obvious agent of confusion. “But there are genes called Snail,” she points out, “and the genes are to do with the development of the mesoderm, so a lot of papers that were about early mesoderm development were being indexed with gastropod snails.” Drysdale wound up writing an exhaustive set of rules that takes into account the presence of terms like malacology, Drosophila, and even zinc.
A screenshot of MAIstro, showing rules for the term snail. Image credit: Public Library of Science
Although the initial thesaurus update went live in December 2012, nine months after the project with AI began, the work of maintaining these rules is never-ending. Drysdale is the only full-time PLOS employee dedicated to keeping the thesaurus up to date, but she gets regular help from various editorial staff, including Kallie Huss from the publishing staff of PLOS ONE, who donates a few hours a week to writing MAIstro rules.
The practice is part careful consideration, part trial and error, and part sheer gut feeling. “You get very good at spotting mistakes, at spotting things that look a little squiffy,” says Drysdale. When a term doesn’t seem to be behaving, she chooses a few key papers and puts them one at a time through a test facility in MAIstro. There, she can rewrite the rules and run the papers past them until they tag appropriately. Before any update to the thesaurus goes live – which happens every six to eight weeks, with a whole host of new rules and rearranged terms – she also runs full-scale searches for terms she isn’t sure about, on a test server that contains every article published in a PLOS journal. “If you’re going to fiddle with the rule for obesity, for example, or cancer, you need to test corpus-wide before you put it out to the public,” she says. “So as I’m modifying rules, based on my experience and my general cautiousness, I flag up ones that I’m particularly worried about.”
“It’s not possible to test everything,” she adds. “So you do your best and try not to worry too much.”
Drysdale gets plenty of input on which terms need repair, from editors, contributors, and readers alike. Two members of the Product Development team, Patrick Polischuk and Sebastian Toomey, put a lot of work into adding new ways for readers to report poorly-targeted terms. When readers see the subject area terms attached to an article – which appear prominently on the right-hand side of the screen next to every paper – they can click a gray flag next to any term to mark it as inappropriate. Drysdale sees a weekly report of every flag that’s been clicked, and can prioritize her work based on those responses. Readers who subscribe to email updates or RSS feeds based on subject area are also given an email address in every notification, to write to PLOS if they get any mismatched article notices.
“People in the project aren’t shy about pointing out when there are mistakes,” says Drysdale, adding that in fact she’s very grateful for readers’ enthusiasm in helping the taxonomy keep up with the human brain. “We always write back and say thank you.”
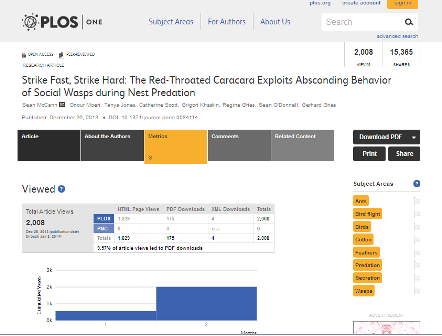
A screenshot of a PLOS ONE article, showing various metadata: the subject terms on the right, plus viewing metrics, and the authors and dates. Reproduced with permission from PLOS
While PLOS has put a lot of effort into revamping its taxonomy over the past two years, reader awareness has lagged behind. PLOS readers have plenty of opportunities to interact with the thesaurus beyond doing searches. With one click, they can sign up for email or RSS alerts associated with any search performed in PLOS. The pages that result from these searches are almost identical to the PLOS ONE homepage – a function that Jonas Dupuich, PLOS Product Manager, likens to having a customized scientific journal. “We’ve essentially made a virtual journal for each of our thousands of subject areas,” Dupuich told Bio-IT World, “and it’s as if you get a homepage where we show recent papers within that subject area. And that’s for every term in our thesaurus.”
The thesaurus can also be accessed through the PLOS API at
api.plos.org, where anyone can create new tools that mine information from the journals. The API works with not only the PLOS taxonomy, but any other piece of metadata tagged to articles, including the authors, editors, submission and publication dates, and metrics like views and citations. With this information, a user could easily graph out, say, the most cited papers in a given subject area, or see how a term has risen in prominence by charting readership of its associated articles over time.
“We have yet to catch up with the promotion and integration of a lot of these features,” says Dupuich. “But by implementing them without the promotion, it’s allowed us to refine and make sure they’re working in the way we want.”
PLOS also uses its thesaurus metadata on the back end, during the publication process. Submitted articles get tagged before peer review, helping the editorial board select the best editors. Contributors to PLOS can even tag themselves with thesaurus terms, to indicate what kinds of papers they’d like to edit. “So we have the areas of expertise of the various folks who interact with our system,” says Dupuich. “Not only do we have the papers classified, but we have people classified as well.” These tools will evolve further in the future; in particular, Dupuich wants to use machine intelligence to choose peer reviewers for papers as well as editors.
This kind of careful attention to article metadata is an essential part of the Public Library of Science’s open access project. Their goal is not just to help academics pad their CVs with peer reviewed publications, but to make sure high-quality research can reach its audience in the scientific community. A surprising amount of work goes into making sure that, when a reader searches for cell growth, or subscribes to updates about tissue engineering, they’re not forced to slog through articles about prison cells and bridges. “It’s a unique problem for PLOS to solve,” says Dupuich, “because we publish in such broad subject areas – for us to provide as much guidance as possible to research that is relevant to the reader.”