Function Follows Form: A New Look at Genome Folding
December 19, 2014
By Allison Proffitt
December 19, 2014 | Fitting a two-meter strand of DNA into a nucleus a few microns long is no simple thing. The genome isn’t wadded up and stuffed into every cell in the body, it’s folded meticulously. A five-year effort to look at the genome inside cells suggests that these folds may play crucial roles in function.
The result is of the work is a “gene-resolution map of the human genome in 3D” published last week in Cell (DOI: http://dx.doi.org/10.1016/j.cell.2014.11.021), said co-first author Miriam Huntley, a Ph.D. student in the Harvard School of Engineering and Applied Sciences.
Huntley and co-first author Suhas Rao, a researcher at the Center for Genome Architecture at Baylor, looked at the architecture of the nucleus in a new way, which gave them a clearer view of what was going on.
They used a new version of the Hi-C protocol, originally published by Erez Lieberman Aiden in 2009, to comprehensively map chromatin contacts genome-wide (see, New Technology Reveals Genome’s Shape). Aiden, senior author on the new study, is now director of the Center for Genome Architecture at Baylor College of Medicine and an assistant professor at Rice University.
The Hi-C protocol combines DNA proximity ligation with high-throughput sequencing in a genome-wide fashion. “The Hi-C protocol works by gluing the genome together, cutting it into millions of pieces, and taking these blobs that have pieces that came into close proximity but didn’t necessarily come from the same place in the [linear] genome,” Rao said.
In the original protocol, the genome was taken out of the nucleus before it was imaged. The new protocol—in situ Hi-C—leaves it in the nucleus.
“We realized there were so many things in the original protocol that were shaking up the nucleus, that were shaking up the chromatin, shaking up the DNA. A huge amount of effort here was figuring out how we can do this that perturbs the nucleus, perturbs the nuclear chromatin, the least,” said Aiden. “That’s where the logic comes from, let’s do this in the intact nucleus… let’s try to avoid disturbing the structure.”
The new technique resulted in far more data than before. Hi-C maps created with the new protocol comprise over 5 Tb of sequence data recording over 15 billion distinct contacts, an order of magnitude larger than all published Hi-C data sets combined.
The new protocol is akin to taking pictures with a better camera, Rao said. Using the new maps, the researchers were able to clearly discern domain structure, compartmentalization, and thousands of chromatin loops.
That the genome folds into loops is not news, said Rao. “People have been looking for these loops since the ’80s,” he said. “In the ’80s people started to realize that there were these sequences that were far away from genes, but they kind of controlled the expression of genes. A kind of looming question was, ‘How do these things talk to each other?’”
Early on, loops were thought to be involved. “The expectation was that you’d see millions of loops just because there are so many regulatory elements on top of a gene,” Rao said, “but in actuality what we found by looking at high resolution data is that… there are only 10,000.”
The loops vary by cell type; different folds yield different function. Rao and Huntley and their teams tested nine different cell types and found that 30% of the loops link gene promoters to distal regulatory sequences.
“If a gene at the beginning of a loop—if the pinching is happening at the beginning of a gene—that’s often associated with that gene turning on,” said Huntley. “If you have different loops forming in different cells, that means different genes are getting turned on in different cells. This provides the beginning of an explanation for how different cells are achieving different functions, how different tissue types are achieving differing functions.”
The groups found a series of common folds repeated throughout the genome. Loops were found throughout the genomes of both humans and mice, though locations differed from cell type to cell type. “Domains” describe the way the genome within the loop folds and condenses, and “subcompartments” describe how different domains pack together.
(Highlighting the similarities to origami artistry, the researchers created a video to illustrate how common folds in the genome can create near-endless variety.)
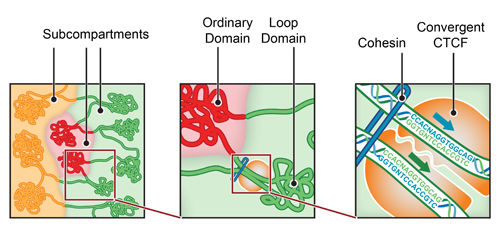
In every case, bits of the genome that would be very far apart if the genome were stretched out linearly, end up folded together in a precise manner. That spatial organization suggests new ways that the genome works.
Crucial to the folding seems to be a single protein called CTCF. CTCF is known to be involved in the regulation of the 3D structure of chromatin, and loops are almost always associated with CTCF. In order for a loop to form, CTCF elements must be pointing toward each other.
But why CTCF binds to form some loops in one cell type and different loops in another cell type isn’t yet clear.
“Why CTCF will bind in one location versus in another location depending on which cell type is probably related to these other ideas like epigenetics… What other chemical modifications are happening? How are things different in this cell type versus the other cell type?” explained Huntley.
Aiden said, “CTCF… is part of a larger complex that works in somewhat mysterious ways. What’s clear is that this complex—of which CTCF is the business end—is sort of like a staple. It glues bits of chromatin together. But the exact mechanism of that stapling is unknown and mysterious.”
“In biology, really big advances have been made every time we’ve been able to figure out the structure of something,” said Rao. “With the genome, there was a lot of work to be done before we could even start thinking about how this is packaged up… But now, we’re kind of at the point where we can start to think about how all of these pieces fit together. How do these things interact? What are the players? I think we’re at a tipping point for really understanding how the genome works, and the folding is essential to that.”


