Citizen Science in the Unexplored Terrain of the Brain
By Aaron Krol
May 13, 2015 | The game is simple. On the left-hand side of the screen you see a cube containing a misshapen 3D figure, a bit like a tree branch with a gall infestation. To the right is a razor-thin cross-section of the cube, a grainy image of overlapping gray blobs. Clicking on a blob colors it in, like using the paint bucket tool in MS Paint, while also sending colorful extensions out from the branch to the left. Working your way through 256 of these cross-sections, your job is to extend the branch through the cube, identifying which blobs are continuous with the branch and which are nearby distractions.
It hardly sounds like a game at all, but strange to say, there’s something very compelling about playing EyeWire. Maybe it’s watching the branches grow and fork as you discover new connections. Maybe it’s how quickly you can rack up progress, almost not noticing time go by as you span your branches through cube after cube.
“It draws you in,” says Nikitas Serafetinidis ― or Nseraf, as he’s known in-game. “There’s an unexplained component that makes this game highly addictive.”
Serafetinidis is the world record holder in EyeWire, a game whose players are helping to build a three-dimensional map of brain cells in the retina. The images in EyeWire are in fact photos taken with an electron microscope at the Max Planck Institute of Medical Research in Heidelberg: each one represents a tiny sliver of a mouse’s retina, just 20 nanometers thick. The “blobs” are thin slices of closely adjoined neurons, and the “branch” shows the path of a single cell, which can cross through hundreds of thousands of those images.
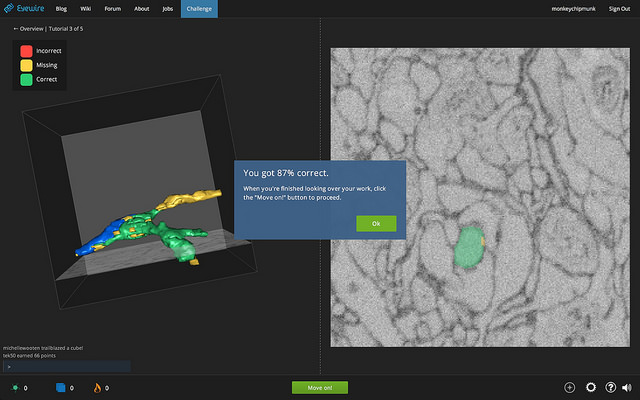
In this screenshot from the EyeWire tutorial, a single extension of the cellular branch on the left passes through the microscope image on the right. Image credit: EyeWire
Piece by tiny piece, players like Serafetinidis have been building up models of cells whose long, stringy dendrites — the tentacle-like extensions that neurons use to communicate — interweave with one another in complex webs. EyeWire relies on the consensus of these players to determine which cell model is most accurate. Top players, like Serafitinidis, can review and finalize the structures for the most difficult cubes.
For Serafitinidis, staying at the top of the EyeWire leaderboard is a serious commitment. While he works from his home in Athens as a 3D animator (his main project is a web game called The Greek Wars, part of an effort to create more homegrown industries in Greece), he cycles back and forth between his animations and EyeWire, usually putting in a couple of hours a day. “There are other days I just wake up and play until I go to sleep,” he adds, a habit he developed while first trying to grab the top score.
Although most EyeWire players have no formal training in neuroscience, their efforts have contributed to one of the world’s most complete resources for exploring the shape and arrangement of neurons in living tissue. They have also been instrumental to an important finding about how the brain detects motion, published last year in Nature (doi: 10.1038/nature13240). Serafitinidis appeared, as Nseraf, in an extended list of coauthors on that paper, along with over 1,500 of his fellow players.
“You sort of feel like you’re part of the greater team,” says Serafetinidis. “There are quite a few regular players that have become friends, and we chat through Facebook and Snapchat.” They also act as mentors to newer players, guiding them on how to interpret difficult images, and as moderators keeping the chat window civil and supportive. In effect, the leading EyeWire players have created and nurtured a huge body of expertise in the art of building neuron maps, far beyond the capabilities of academic scientists.
“We have 180,000 players, and they come from all over the world,” says Amy Robinson, Creative Director of EyeWire. “We know the top players personally, because we spend time in there talking to them. When we build new features we have them beta test them, we ask them for their ideas, we respond rapidly to their bug reports and questions. It’s driven by a quite strong community.”
That community has helped EyeWire expand since its creation in 2012, as a part of the MIT lab of Sebastian Seung. While Seung has since departed for the Princeton Neuroscience Institute, EyeWire remains a semi-autonomous project run by Robinson at MIT, hiring new developers and, increasingly, looking to public volunteers and competitions to contribute new mapping algorithms.
“I think that in years to come we’ll see a lot more of this crowdsourcing, not just for analysis of the data but to build automated pipelines,” says Robinson. “What we’re seeing with EyeWire, and what a lot of these crowdsourcing challenges are seeing, is that you don’t necessarily need to have any background in the field in which you’re working. You just have to be a curious, talented, passionate developer.”
Blind Spots
The information age is changing many of the basic skills needed to conduct cutting-edge science. In the life sciences, vast amounts of data, from new devices like DNA sequencers and wearable sensors, have given rise to a professional class of computational biologists, whose skills at data management are essential to supporting traditional lab work.
Neuroscience has a special problem, because it relies heavily on imaging data — that rare type of data that computers are less skilled at interpreting than people. Microscope images of the brain are the key to classifying neurons into different cell types, and to seeing where synapses form between them. Unfortunately, computers are very poor at this kind of image recognition task.
“Connections are crucial for deciding the functions of cells,” says Jinseop Kim, a postdoctoral researcher at Princeton involved in the EyeWire research. While it may be hard to conceive of high-level brain functions, like spatial memory or visual processing, as products of the physical location of individual neurons, something as simple as the distance between two cells can often prove essential to their behavior.
When Seung’s team started working on the retina, they wanted to find a new approach to interpreting images from this neuron-rich part of the eye, one that could overcome computers’ failure to reliably track cells in three-dimensional space. Human observers could outperform any artificial intelligence system, but the process of coloring in images by hand was too slow to be practical. While a mouse’s retina is a very small piece of tissue — not much more than a centimeter square and less than a tenth of a millimeter thick — within that tiny region of the eye are one and a half million neurons. By 2012, Seung realized that making even a small dent in that structure would demand far more manual work than the lab could ever perform on its own.
“At that time, crowdsourcing was a hot issue,” Kim says. “By the time we started developing EyeWire, Foldit was very successful.” Foldit, which enlists players to figure out how strings of amino acids fold into proteins, was the first game to take off as a way to recruit thousands of “citizen scientists” to solve biological problems.
“Foldit showed that you can take a complicated, difficult task, and you can gamify it and get people to play,” says Robinson. The Seung lab chose to follow Foldit’s example, releasing a version of their neuron-mapping software to the public and letting volunteer players fill in the gaps where artificial intelligence failed to complete cellular branches.
From the beginning, EyeWire took its crowdsourcing ethos seriously, inviting anyone with good ideas to contribute. Robinson herself, then working as a TED events organizer, benefited from EyeWire’s openness to non-neuroscientists’ contributions: she was asked to join the project after writing a letter to Seung as a “crowdsourcing expert” with ideas to spur more player engagement.
Under Robinson, EyeWire has added team competitions, new “badges” for top players that give them special abilities like culling incorrect cubes, and big promotions around solving some of the hardest cubes. “We have looked a lot at games like Angry Birds, these viral online games where people are spending billions of hours collectively just solving puzzles,” Robinson says. The game is constantly being tweaked for better performance and more of the elusive “stickiness” that keeps players coming back.
Today, the EyeWire community maps roughly one cell a day — an impressive feat, since each neuron can span as many as 3,000 cubes as its dendrites stretch out in all directions. Serafetinidis alone has completed almost 150,000 cubes, the equivalent of perhaps 100 whole neurons.
Form and Function
Three-dimensional pictures of brain cells contribute more than cool slides for conference talks. Knowing which cells are present in a system, and where they connect, is the first step to learning how their interactions give rise to mental functions.
“Cell classification is not a simple task,” says Kim. “We don't have a complete knowledge of cell types. Not yet.”
One question the Seung lab has pondered is how the brain processes motion. It was already known before the lab started its work on the retina that certain neurons, called starburst amacrine cells, respond strongly to motion in a specific direction. These cells are activated when photoreceptors in the eye pick up changes in lighting conditions ― say, a passing car causing a sudden decrease in light.
But starburst cells don’t react to just any lighting signal. Instead, a starburst cell fires only when a signal begins at the center of its body, and moves out across its dendrites. Combining messages from many of these cells, the brain can reconstruct which way an object is moving, but how the starburst cells could tell that a motion was “outward” rather than “inward” was a mystery.
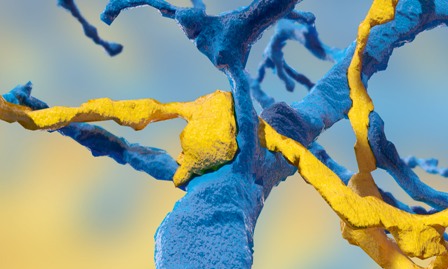
EyeWire’s data was perfect for settling this question. Starburst amacrine cells, as their name implies, have a very distinctive shape, like a firework going off, which makes them easy to spot in 3D maps. The EyeWire community has built several complete maps of starburst amacrine cells, as well as the surrounding bipolar cells, which respond more directly to light.
In 2014, Seung, Kim, and their colleagues published a paper in Nature showing how the arrangement of those bipolar cells could tell a starburst cell when an object was moving “out.” The answer has to do with the timing of light responses: one type of bipolar cell fires almost as soon as it experiences a change in lighting conditions, while a second type waits for up to 100 milliseconds before sending out a signal. The two types are visually different enough to tell apart in an EyeWire reconstruction.
Looking at the EyeWire maps, the researchers realized that the faster bipolar cells connect with the farthest branches of starburst amacrine cells, while the slower types connect closer to the center. That means if a motion begins at the center of a starburst cell, and continues out to the edge, signals from both bipolar cells will reach the starburst cell’s center at almost the same time, creating a critical mass that causes the starburst cell itself to fire. Motion in any other direction will only result in isolated signals to the starburst cell.

This reconstruction of a neural cell carpet, featuring starburst amacrine cells, bipolar cells, and ganglion cells, hints at the huge complexity of the networks EyeWire players are helping to map. Image credit: EyeWire
As data from EyeWire continues to build, the Seung lab hopes to make more discoveries about how the retina functions. The players, too, often end up learning a little more about the workings of the brain, through communications with the Seung lab, a dedicated EyeWire wiki, or just by exploring the shapes of the neurons they painstakingly reconstruct.
Ultimately, however, the project’s leaders see EyeWire as a first, temporary step to harnessing the power of the crowd for a much deeper expedition into the brain. “We like to think of EyeWire as the proof of concept,” says Robinson, “but in years to come we’ll be working with other labs around the world to help them analyze their data with the crowd, and help them crowdsource AI, and also to help them communicate and share their research.”
The next step is to move beyond the retina — which, with relatively few and well-known cell types, is a simpler model for neuroscience — to the brain itself. Seung is interested in exploring the piriform cortex, a region near the brainstem involved in the sense of smell, which will be the focus of the first successor program to EyeWire. That will take entirely new artificial intelligence, which could be built by a combination of EyeWire employees and outside contributors.
The cortex presents harder problems than the retina, for both computer intelligence and human players. Different staining techniques, which reveal the structures inside of cells, are used for microscope images in this region, making it even more difficult to tell where one neuron ends and the next begins. There’s also a far greater variety of cell types deep in the brain, many of which are still unknown to science.
“From a scientific point of view, we have less knowledge of the piriform cortex,” says Kim. “In the retina, we have pretty abundant knowledge on cell types, and their connectivity, and cell types in the retina have very common structures. But cells in the cortex are very complex, way more complex than cells in the retina. It’s harder to get an accurate reconstruction, and it’s harder to make a correct map of the connections.”
Even in the familiar ground of EyeWire, the brain continues to offer surprises. “There are many cells we’ve mapped that we’ve never seen anything like,” says Robinson. “That no one’s ever seen anything like.” That’s both the challenge and the value of using citizen science to map the brain: in this little-explored terrain, no one really knows what we’ll find when scientists finally have the resources to rebuild neural circuits and look at neurons in their natural environment.
“If you look at the animations and images that come out of EyeWire, you’re seeing real, nanoscale resolution, accurate models of cells,” Robinson adds. “A lot of people have a curiosity and interest in the brain, but they’ve never actually seen it.”
You can play EyeWire at https://eyewire.org/signup, or learn more about the project at http://blog.eyewire.org/en/.


