The Sequencing App and the Quest for Fun
By Allison Proffitt
September 26, 2016 | The latest way to get into your genome is a $50 app-based venture that promises to give you a peek into your ancestry and tell you what kinds of bacteria you’re harboring in your mouth.
Tasty.
Earlier this month Joe Pickrell and his lab formally launched Seeq, an app that offers ultra-low coverage sequencing of the genome, oral microbiome sequencing, and ancestry reports. The goal is an extremely inexpensive infrastructure for people to get a little bit of genomic data and participate in research in a way that’s easy and fun.
Pickrell, a professor of biology at Columbia and the New York Genome Center, has been working on Seeq for more than a year. “We think people seem quite interested in participating in genetics research, but primarily not in the standard sort of mode where we collect a lot of data from you and then tell you, ‘Thanks! Have a good day.’ People seem quite interested in getting something back,” he told Bio-IT World.
So of course they started with an app.
With a small lab of only five, Pickrell knew he’d need to design the Seeq process carefully, so from the beginning the platform was built for mobile engagement. Users download the free app and set up a simple account with an email address and password. Seeq returns two types of data to users: ancestry data and mouth microbiome data. You can start by exploring sample ancestry and mouth microbiome datasets from “Chuck Darwin”, whose last meal included peaches, and the SeqBot, who has three kinds of viruses in his (her?) mouth, and genetic signatures from five regions of the world.
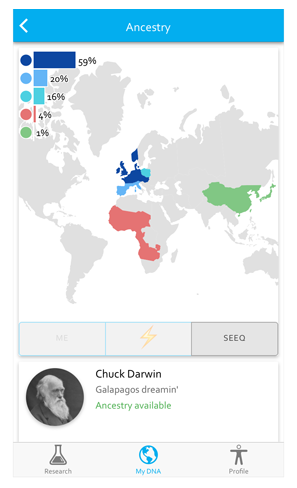 If that looks good, users can click through a five-screen consent form, and participate in research by completing phenotype questionnaires on personality, sleep patterns and circadian rhythms, facial features, quirks about your body, and mouth hygiene.
If that looks good, users can click through a five-screen consent form, and participate in research by completing phenotype questionnaires on personality, sleep patterns and circadian rhythms, facial features, quirks about your body, and mouth hygiene.
All of this happens before the first splatter of spittle lands in a tube. If a user is intrigued, then he or she can contribute $50 to cover shipping and receiving and start the collection process.
Low Coverage, High Value
Seeq uses the same saliva collection tubes as other direct-to-consumer sequencing companies, but the sequencing itself is different.
Pickrell’s team is using ultra-low coverage sequencing technology to impute each individual’s genome. While clinical sequencing is often done at up to 60x coverage, ultra-low coverage sequencing is closer to 0.1–0.5× coverage of the genome. Of course that means there’s a lot that is missed, but maybe not as much as you’d think.
In a 2012 Nature Genetics paper (doi:10.1038/ng.2283), Bogdan Pasaniuc and his colleagues showed that extremely low-coverage sequencing captures almost as much of the common and low-frequency variation across the genome as SNP arrays. Pasaniuc et al. demonstrated that, in principle, “association statistics obtained using extremely low-coverage sequencing data attain similar P values at known associated variants as data from genotyping arrays, without an excess of false positives.”
“We see that in practice, this works,” Pickrell explains. “Effectively we’re taking a random 10% of your genome and sequencing it once... We’re getting 10% of your variation.”
The results aren’t going to be terribly useful for any one person. This isn’t a clinical or medical tests, and these aren’t results you’ll take to your doctor to inform healthcare decisions. But if the Seeq team combines such results from many people, Pickrell believes there’s much to be learned.
“In aggregate, if we have 10,000 people… we’ll be able to see the right statistical association and discover the right genetic variants, even if we can’t tell you that you personally have [a condition.]”
It’s a genome-wide association study, of course, and as sequencing gets cheaper and more clinical, GWAS studies seem to be falling out of favor. But Pickrell sees great value in GWAS studies.
“It’s all a tradeoff. If you had a fixed amount of money, would have do a genome-wide association study of 10,000 people of a whole genome sequencing study of 100 or 1,000 people? What we really care about is sample size. We want to get the sample size as large as possible, and so we’re willing to make those tradeoffs in terms of the clinical quality.”
Getting large sample sizes means engaging users and keeping the service inexpensive. Doing low coverage sequencing helps Seeq keep costs down, but the sequencing isn’t the only cost.
“I’m a statistical geneticist, and I naïvely thought, ‘Great, we’ll just sequence less and that will be lower cost.’ But it turns out there’s this fixed cost associated with the sample, which is the DNA extraction, the library prep, the shipping, the handling, all of that,” Pickrell admits. “We spent a lot of time trying to get that right and get it as inexpensive as possible.”
The Seeq team streamlined DNA extraction and library prep—Pickrell said those processes will be released publicly soon—and is relying heavily on robotics in the lab. Even with only five staff members, he said the lab is, “nowhere near capacity yet.”
“We want the thousands of library preps to be done by a robot, not a person. We want the person to be thinking about ways to improve the library prep.”
Cabinet of Curiosities
With an affordable pipeline in place, the next step is making it fun.
Especially in new research areas and for things Pickrell calls “curiosities”, genome-wide association studies are a great way to lay the foundation for future research without needing significant funding, he believes.
Though Seeq users aren’t getting reports on medically actionable health findings, they will get data back on their ancestry and—interestingly—their mouth microbiome. They can also pick some of the curiosities that the Pickrell lab will dig into.
“We want it to be not just us dictating what we want to do, but people telling us what they’re interested in doing with their data, and what they’re interested in learning,” he said.
In his blog announcing Seeq, Pickrell suggested that users might ask questions about familial traits or idiosyncrasies. “Do both you and your father have inexplicable cravings for pickle juice that you suspect might be genetic?” Pickrell writes. “Let us know, and we’ll see if we can identify the gene (or genes).”
But even a question about pickle juice can reveal some interesting medical findings in a GWAS setting.
“The pickle juice example comes from studies of hypotension, or low blood pressure,” Pickrell said. People with very low blood pressure tend to crave salt… and pickle juice is loaded with salt. So there’s this association between low blood pressure and cravings for pickle juice.”
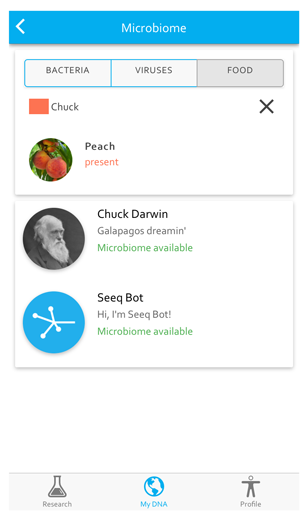 To dig deeper, Seeq plans to regularly send out mini-questionnaires to users via the app that include questions driven by both the community and the Seeq team.
To dig deeper, Seeq plans to regularly send out mini-questionnaires to users via the app that include questions driven by both the community and the Seeq team.
“Effectively what we’re doing is research on the genetics of low blood pressure but through this proxy, which is more interesting and more engaging.”
Engaging the consumer is a main point of the Seeq mission. The Seeq customer is curious, but probably hasn’t yet gotten involved in direct-to-consumer sequencing, Pickrell believes. “I know lots of people who are curious about genetics but it’s too expensive and it’s kind of scary. Our target audience is those people.”
One of the first industry movers, 23andMe, prices its health, ancestry and traits report at $199; its ancestry-only option is $99. AncestryDNA also runs $99. That may be too expensive for some. “About 50% of people probably don’t want to learn if they are at risk for some disease they can’t do much about,” Pickrell posits. “I, on the other hand, want to know everything, but I know many of my friends and family members would prefer not to know things like that.”
The most challenging findings from a Seeq test would be surprises in ancestry, Pickrell believes. It’s a point highlighted in the five-screen consent process on the Seeq app. “That’s probably the number one thing that we’re thinking people might learn that’s unexpected and might upset them.”


