Next-Generation Knowledgebases Are Required For Interpreting Data In Precision Oncology
By Susan M. Mockus
September 10, 2018 | Bio-IT World Best Practices Award Honorable Mention | In May, we announced the winners of the Bio-IT World Best Practices Awards. Each year these awards are given with the goal of celebrating projects that highlight excellence in bioinformatics, basic and clinical research, and IT frameworks for biology and drug discovery. While only five projects are rewarded, we can’t help but recognize the remaining finalists and honorable mentions for their efforts in elevating the critical role of information technology in modern biomedical research. Here is one of the two honorable mentions. – The Editors
Advances in precision oncology have created a demand for scalable interpretative tools that address the growing complexity of genetic/genomic data and corresponding treatment modalities. Patient clinical cases where a single mutation may predict therapeutic response are often an exception to more complicated molecular scenarios that transverse a wide array of alterations including multiple mutations, aberrant expression, copy number variations, fusions, tumor mutational burden, and microsatellite instability. On the treatment modalities side, acquired resistance and targeted therapies in combination with immunotherapies have further made navigating the terrain incredibly challenging.
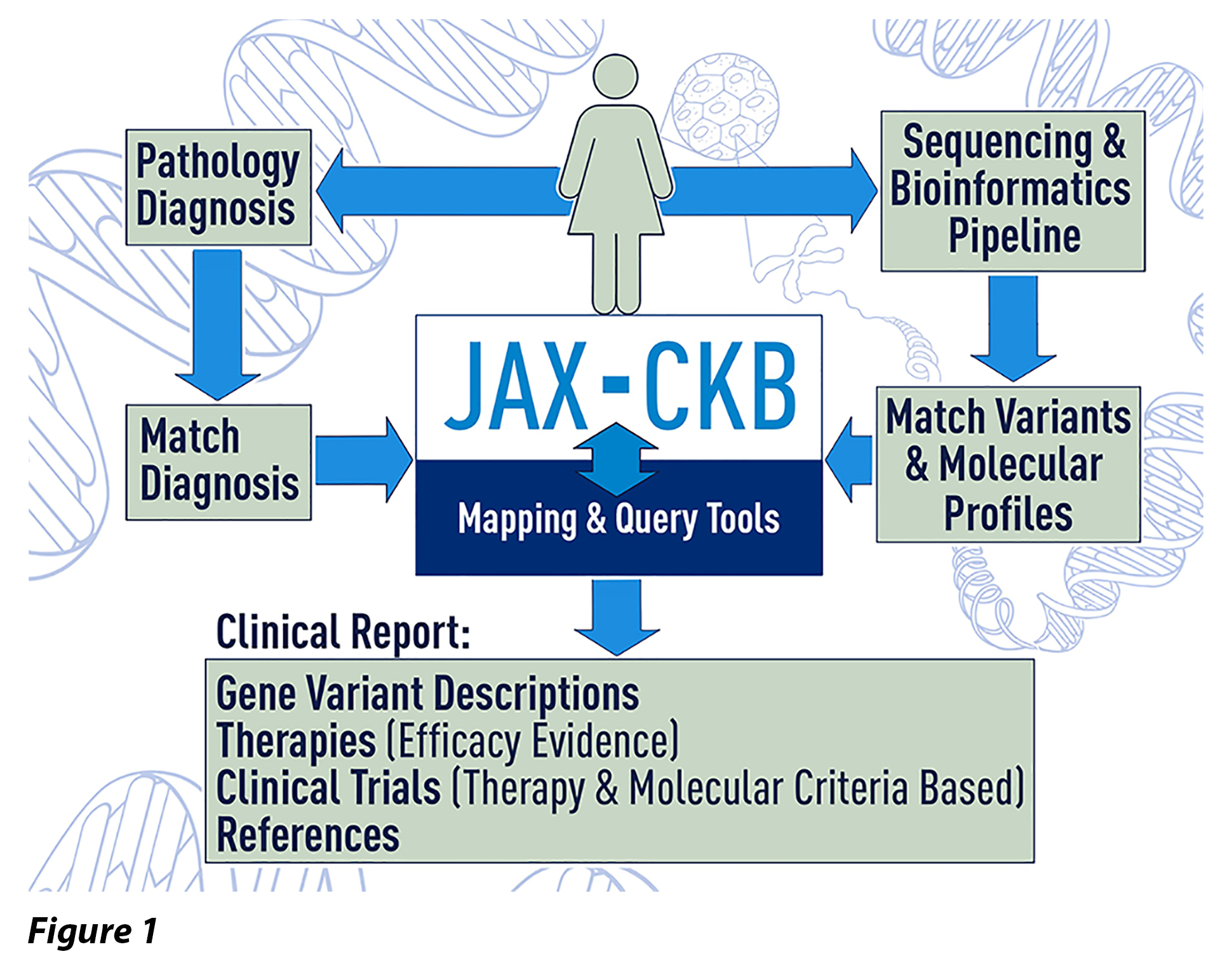
While precision medicine has undeniably had an enormous impact on oncology and the understanding and treatment of cancer patients has improved, data interpretation still remains a significant bottleneck. To address this, The Jackson Laboratory (JAX) built a Clinical Knowledgebase (JAX-CKB) to provide evidence-based information to clinicians, researchers, and ultimately patients. Initially developed to support JAX’s in-house Clinical Genomics Laboratory, JAX-CKB has evolved to be a dynamic and digital encyclopedia for precision oncology. JAX-CKB is an expertly curated and publicly accessible relational knowledgebase of gene variants, targeted therapies, efficacy evidence and clinical trials related to cancer.
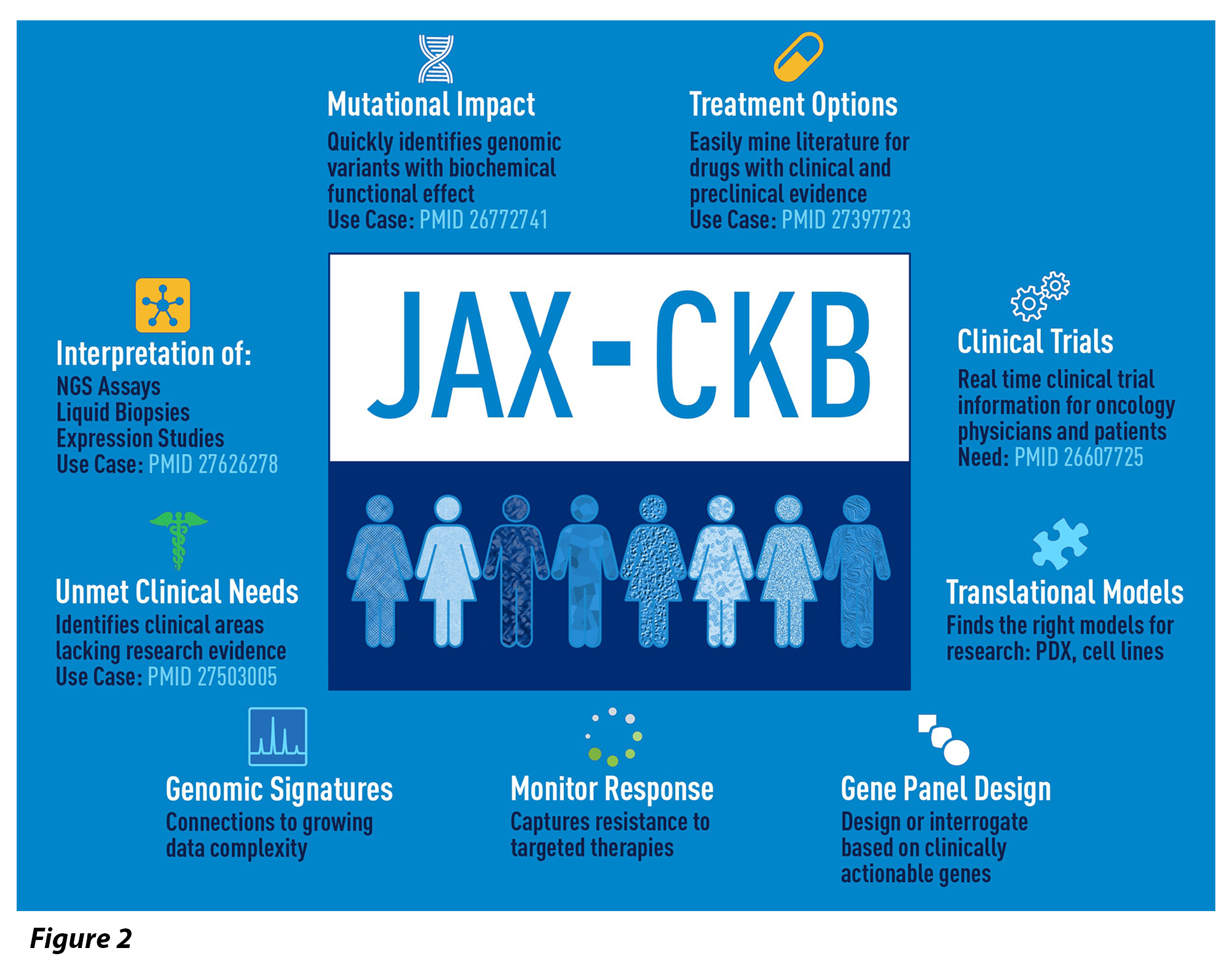
JAX-CKB has over 30,000 users and continues to inform patient cancer care while contributing to technology and methodology development. Key commercial collaborators are aiding JAX-CKB sustainability by leveraging machine-learning capabilities and third-party software platforms have integrated JAX-CKB to enable easy and scalable accessibility for informing cancer patient care.
Precision Oncology Data Interpretation Needs
A fundamental challenge to precision medicine is the ability to easily connect a patient’s genetic/genomic variants with a therapeutic approach, which could include either FDA-approved targeted therapies or targeted therapies in recruiting clinical trials. Recent studies have demonstrated that linking patients to relevant targeted therapies based on genomic data has the potential to achieve higher clinical success relative to recruitment solely on histological subtype.
Leveraging the power of an integrated knowledgebase, we can identify opportunities for clinical intervention, including available clinical trials for targeted therapies. The level of evidence for any given molecular profile and therapeutic response can be further characterized using the AMP/CAP/ASCO somatic guidelines by mapping JAX-CKB controlled vocabularies to this tier ranking system.
A comprehensive knowledgebase can also provide a unique view into the landscape of molecular targets and therapies, uncovering potential opportunities for new research and development. In addition to supporting clinical reporting, the JAX-CKB allows visibility into deficiencies in the characterization of potentially actionable mutations and therapeutic interventions, exposing opportunities for research that have the potential to advance cancer treatment.
Simplified Mechanisms Of Data Capture And Retrieval Is Required For Complex Cancer Data
Since the inception of JAX-CKB in 2014, data have been captured in the context of molecular profiles to address the complexity of cancer. It is well known that more than one molecular change may influence pathway response to targeted therapies and this aspect of complex molecular profiles enables accurate representation of the underlying biology. The complex profiles can be further leveraged using enhanced searching to predict patient response to a combination of targeted therapies and/or immunotherapies. Furthermore, these complex molecular profiles can identify mechanisms of emerging drug resistance by comparing differing responses that are tissue dependent.
Additional content to supplement gaps in understanding of cancer molecular profiles will include primary data from JAX’s patient-derived xenograft (PDX) models, in which well-controlled studies of targeted therapy response will be captured. Another existing gap in precision oncology relates to therapies and identification of the genes they target. This deficit has been addressed in JAX-CKB by developing a comprehensive list of therapies (2,398 as of 8/7/17) curated to 266 drug classes, internally designed to capture gene targets. The drug class list is currently a flat list of controlled vocabularies, with plans for future conversion to a navigable drug class ontology for non-restricted use by the medical and scientific communities.
Machine-Learning Assisted Curation Ensures Scalability
The success of precision cancer treatment hinges on efficient access to comprehensive and up-to-date actionable clinical knowledge. Such unstructured knowledge is scattered in tens of millions of publications, with thousands added daily. Structuring knowledge through controlled vocabularies and ontologies followed by text-mining and machine learning technologies, ensures a scalable knowledgebase. Machine-reading technology can potentially alleviate the data volume problem by automatically extracting candidate facts from text. Expert curators can then quickly inspect the results and triage publications for inclusion through an assisted curation tool.
Real World Use Case
Approximately 9,000 new cancer cases occur each year in Maine. Oncologists and other healthcare providers often struggle with identifying optimal therapies for many of these patients using conventional diagnostic methods and clinical guidelines. The Maine Cancer Genomics Initiative (MCGI), enabled through generous financial support from the Harold Alfond Foundation, leverages the strengths of key medical and bioscience research institutions in Maine to create an alliance focused on precision cancer diagnostics and treatment.
Over the course of three years, 1,800 Maine cancer patients will have molecular tumor tests at no cost in an effort to find more effective cancer treatments with reduced side effects. The initiative will also work with clinicians and cancer patients at every cancer care center in Maine to understand how personalized medicine can potentially improve their outcomes. JAX-CKB is at the heart of this effort, powering the content of the clinical reports for each patient, and helping clinicians and tumor boards understand the molecular characteristics of each patient’s tumor, and explore optimal therapies and clinical trials of potential interest to the patient.
Undertaking and maintaining a clinical knowledgebase is no trivial task. Modular and flexible designs are required to allow adaptability to the ever-evolving field of precision oncology. Combining the power of machines with the human mind will ensure scalability to meet the complex data interpretation needs in precision oncology. JAX-CKB, created as a dynamic and digital encyclopedia with sophisticated search capabilities, is a next-generation knowledgebase serving the translational research and clinical oncology communities.
Dr. Mockus is the Associate Director of Clinical Genomic Market Development at The Jackson Laboratory for Genomic Medicine. She oversees the JAX Clinical Knowledgebase (JAX-CKB) and focuses on innovative product development in molecular diagnostics. She received her Ph.D. from Wake Forest University and her MBA from the Yale University, School of Management. She can be reached at susan.mockus@jax.org.


